shRNA Quality Control
Functional Validation
OriGene has tested many of its shRNA constructs to validate our methodology for selecting ideal construct sequences to include in our shRNA collection. As an example, we have demonstrated the down regulation of endongenous TP53 expression by our shRNA plasmids against TP53 (below) and down regulation of exogeneously expressed HIF1A as well as for other genes [CDC2, CHUK, KIT, MEKK5, PIM1, STAT1].
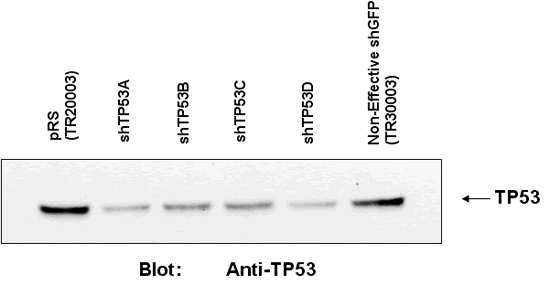
HEK293T cells were plated at 2 X 105 per ml for 2ml into a 6-well plate one day before transfection. One ug Hush plasmids were used together with 4 ul TurboFectin 8.0 for transient transfection as suggested by OriGene transfection protocol. Seventy two hrs post transfection cells were lysed. Five ug protein were loaded on Invitrogen NuPage Gel system and separated proteins were immunoblotted with anti-TP53 () antibody.
Sequence Validation
In order to provide its customers with the highest quality products, OriGene uses both sequencing and restriction enzyme digestion for quality control purposes.
All HuSH plasmid products with shRNA expression cassettes have been isolated using mini-prep DNA purification and have been sequence verified using the primer 5'-TTGAGATGCATGCTTTGCATAC-3' . The sequences are trimmed and matched against the target genes through BLAST analysis. OriGene includes several DNA relaxation agents in the sequencing reaction in order to sequence through the shRNA hairpin structure.
OriGene also uses restriction enzyme digestion for verification of the shRNA cassette cloned into the pRS vector. This is particularly useful for customers who are not able to readily sequence the plasmid. Please see the diagram below for the HuSH shRNA cassette sequence.
HuSH shRNA Cassette Construct (CDC2 29-mer sample sequence)
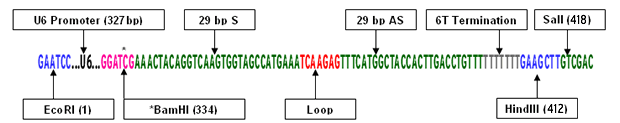
* The original BamHI sequence, GGATCC has been changed to GGATCG during the cloning procedure. HindIII and SalI sites are located at 378 and 384 of the original vector, respectively.
The full vector sequence is
available
.
Verification by Digestion of HuSH shRNA Cassettes and Control Plasmids

* The original BamHI sequence, GGATCC has been changed to GGATCG during the cloning procedure. HindIII and SalI sites are located at 378 and 384 of the original vector, respectively.
The full vector sequence is
available
.
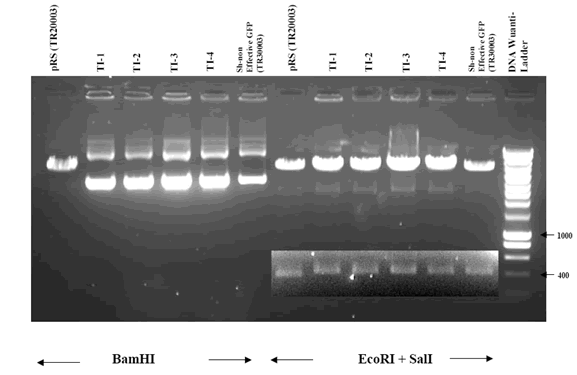
The BamHI restriction site in the pRS vector is altered by the insertion of shRNA cassette. While BamHI digestion will linearize the original vector (TR20003), the successfully cloned HuSH constructs will lose the BamHI site and remain uncut (left panel of the figure below).
Digestion with both EcoRI and SalI will release the shRNA cassette along with the U6 RNA promoter sequence. The EcoRI/SalI or EcoRI/HindIII fragment can be used to shuttle the hairpin shRNA cassette together with its U6 promoter to another vector system for gene down-regulation. EcoRI/SalI digestion will result in a 384bp and a 418bp band for the original vector and the shRNA sequence inserted vector, respectively (right panel of figure below).
The 400bp region was visually enhanced due to the weakness of the original bands after long migration through the gel.